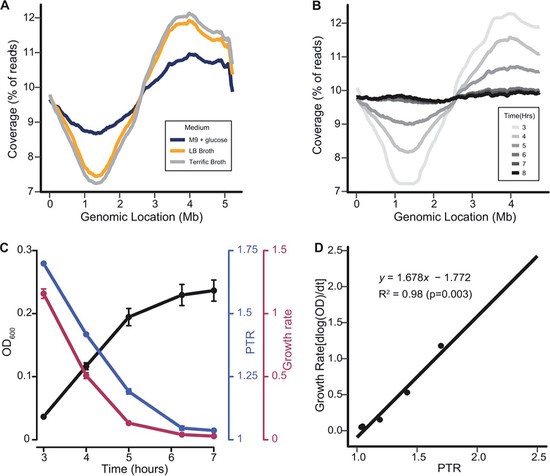
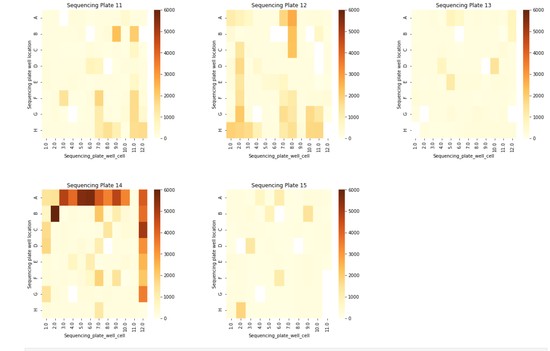
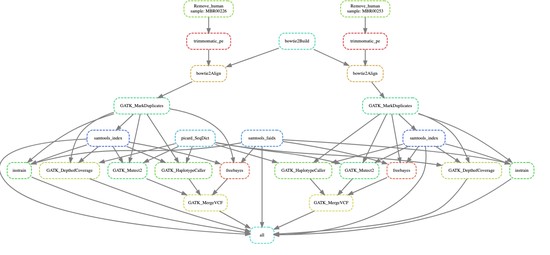
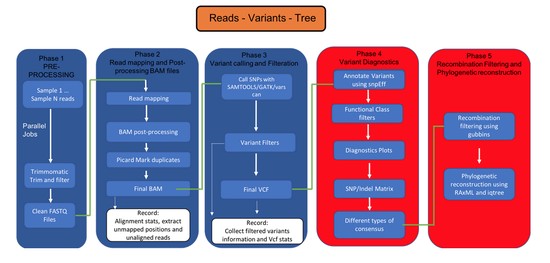
Bioinformatics Scientist with 9+ years of interdisciplinary research experience specializing in the field of Bioinformatics/Computational pipeline development and Multi-omics data analysis.
Interests
- Computational Biology
- Infectious Disease
- Multi-omics and Microbial evolution
- Data Driven Public Health Epidemiology
- Reproducible Research
Education
Master of Science, Bioinformatics, 2014
Georgia Institute of Technology, Atlanta, GA
B. Engineering, Biotechnology, 2011
The Oxford college of Engineering, Bangalore, India